
The revolution in DNA sequencing technologies over the last decade has resulted in an enormous, and ever growing, number of gene sequences, which is doubling every ~18 months. įunding: This work was supported by National Institute of Health The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.Ĭompeting interests: The authors have declared that no competing interests exist. The complete package can be downloaded from the Comprehensive Perl Archive Network at. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are creditedĭata Availability: SmotifTF is a free software package created using Perl and is distributed under the Artistic license version 2.0 (GPL compatible).
Received: MaAccepted: JPublished: August 7, 2015Ĭopyright: © 2015 Vallat et al. Marti-Renom, CNAG - Centre Nacional d’Anàlisi Genòmica and CRG - Centre de Regulació Genòmica, SPAIN Smotif-based modeling is complementary to current prediction methods and provides a promising direction in addressing the structure prediction problem, especially when targeting larger proteins for modeling.Ĭitation: Vallat B, Madrid-Aliste C, Fiser A (2015) Modularity of Protein Folds as a Tool for Template-Free Modeling of Structures. SmotifTF successfully predicts the overall fold of the target proteins in about 50% of the test cases and performs competitively when compared to other state of the art prediction methods, especially when sequence signal to remote homologs is diminishing.
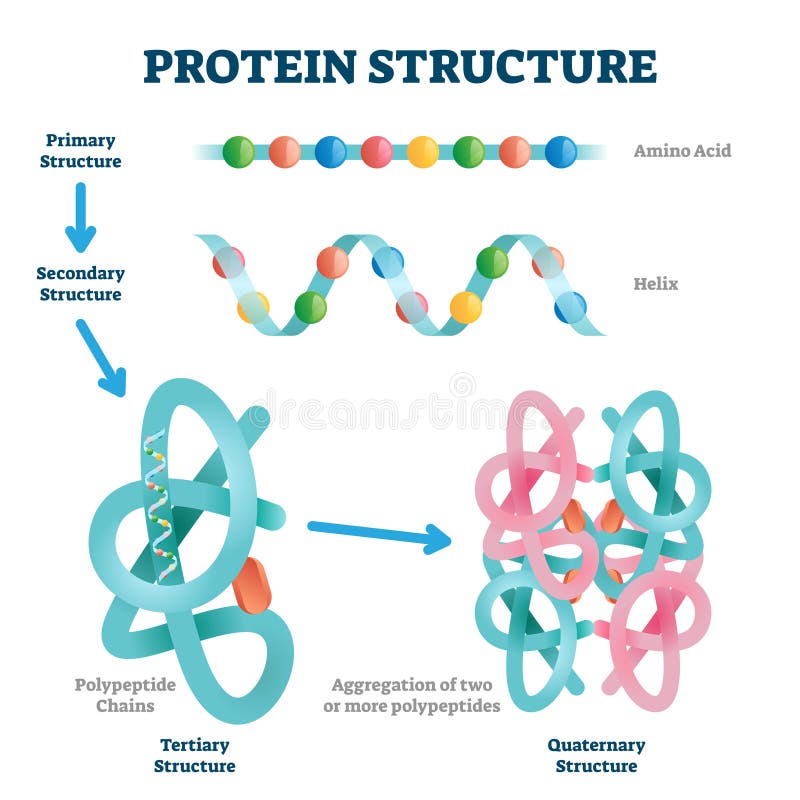
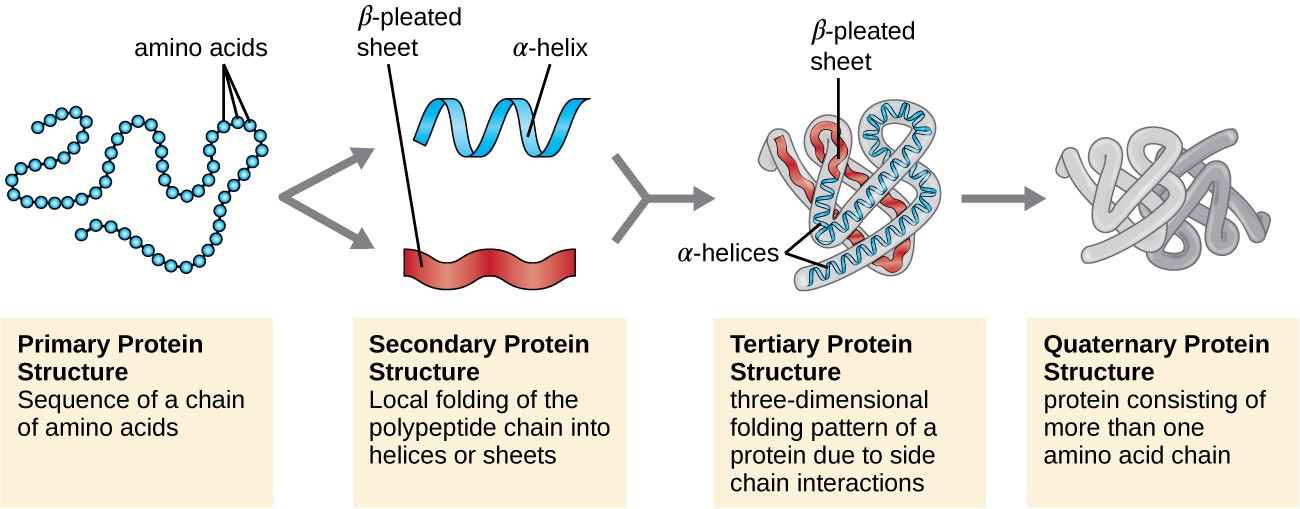
Since the Smotif fragments are larger in size compared to the ones used in other fragment-based methods, the proposed modeling algorithm, SmotifTF, can employ an exhaustive sampling during decoy assembly. This Smotif library is exploited in a fragment assembly protocol to sample decoys, which are assessed by a composite scoring function. The method relies on weak sequence signals from remotely related protein structures to create a library of Smotif fragments specific to the target protein sequence. The library of Smotifs has saturated over time, providing a theoretical foundation for efficient modeling. We present here a novel computational method that uses a library of supersecondary structure fragments, known as Smotifs, to model protein structures. However, template-free modeling methods are much less reliable and are usually applicable for smaller proteins, leaving much space for improvement. While the current structural coverage of proteins is almost exclusively provided by template-based techniques, the modeling of the rest of the protein sequences increasingly require template-free methods. Predicting the three-dimensional structure of proteins from their amino acid sequences remains a challenging problem in molecular biology.


 0 kommentar(er)
0 kommentar(er)
